Not Dead… Yet!
An Introduction to Survival Analysis
Access Presentation
https://www.inqs.info/pres/long_beach/pres_4_22.html
Email: isaac.qs@csuci.edu
Background Information
Background Information
Scenarios
Censoring
Survival Rate
Example
Survival Analysis
Survival Analysis is a set of tools to analyze time-to-event data. This leads to the survival rate which models the time to percentage of data to observe the event.
- To determine the time until a percentile of the data achieves an event
- Determine which factors may affect the survival rate
Scenarios
Background Information
Scenarios
Censoring
Survival Rate
Example
Colon Study
- Colon cancer is a type of cancer found in the large intestine
- Several risk factors affect the survival rate
- We will look at how different medications affect the survival rate
Channel Island Birds
Channel Island Birds
We are interested in how long do bird eggs hatch since being laid
Determine the hatch rate for each species
What happens when there is missing data?
How do we determine the survival rate?
Measure from first recording to event of interest
Construct a probability of surviving up to a certain time point
Data Type
Data is typically recorded as time-to-event data
For biomedical studies, researchers are interested in time from diagnosis to death, known as time-to-death
Censoring
Background Information
Scenarios
Censoring
Survival Rate
Example
Censoring
Censoring is a mechanism where we do not observe the true time-to-event
Not all the time is observed
Three common types of censoring mechanisms: Right, Left, and Interval
CI Birds Example
You design a study where you want to record the time it takes for an egg to hatch for different species of birds.
You spend a week monitoring different bird nests and record times when eggs are laid.
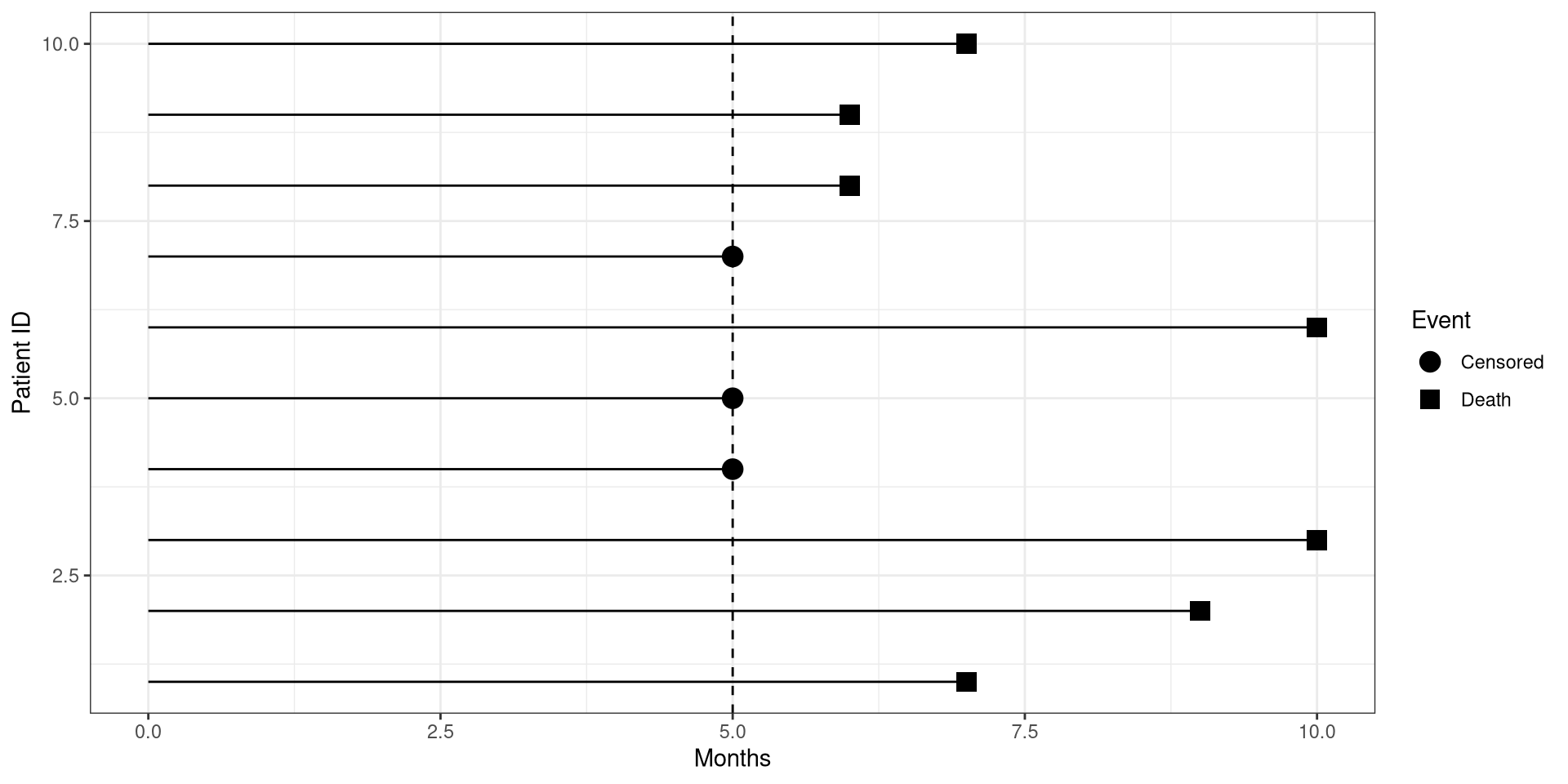
CI Birds Left Censoring
The day before the eggs hatched, your boat broke down and were not able to go to the islands for a week due to repairs.
During this time, 5 eggs hatched!
Left Censoring
library(ggplot2)
dat <- data.frame(ID = 1:10,
t1 = c(7, 9, 10, 5, 5, 10, 5, 6, 6, 7) ,
censored = c(1, 1, 1, 0, 0, 1, 0, 1, 1, 1))
ggplot(dat, aes(x = ID, y = t1, shape = ifelse(censored, "Death", "Censored"))) + geom_point(size = 4) +
geom_linerange(aes(ymin = 0, ymax = t1)) +
geom_hline(yintercept = 5, lty = 2) +
coord_flip() +
scale_shape_manual(name = "Event", values = c(19, 15)) +
# ggtitle("Left Censoring") +
xlab("Patient ID") + ylab("Months") +
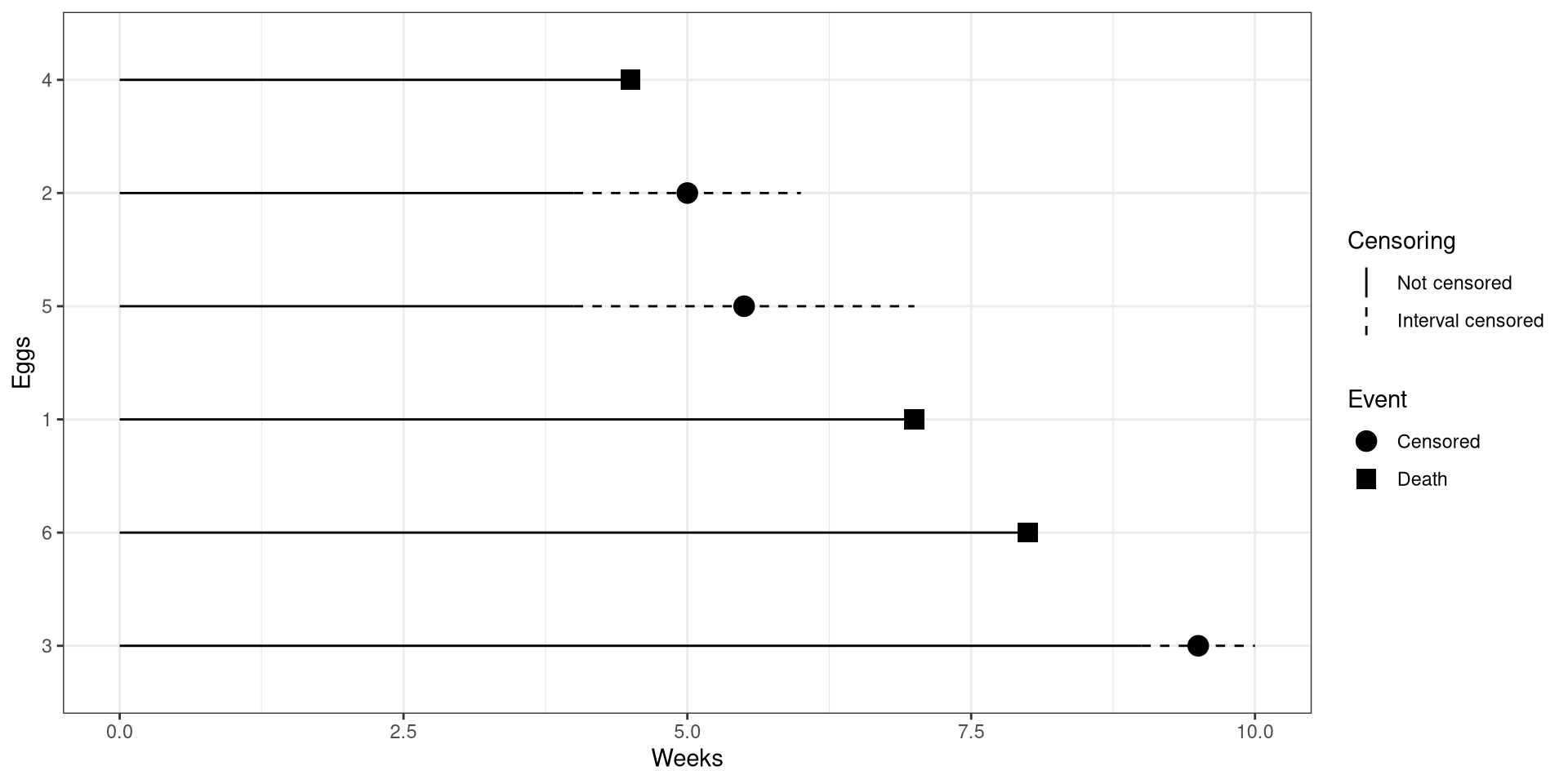
theme_bw()CI Birds Interval Censoring
As you been going out to the islands each day to record whether eggs have hatched or not, you can’t travel to the islands for the next few days due to stormy weather.
During this time, 8 eggs hatched!
Interval Censoring
library(ggplot2)
dat <- structure(list(ID = 1:6,
eventA = c(0L, 1L, 1L, 0L, 1L, 0L),
eventB = c(1L, 0L, 0L, 1L, 0L, 1L),
t1 = c(7, 4, 9, 4.5, 4, 8),
t2 = c(7, 6, 10, 4.5, 7, 8),
censored = c(0, 1, 1, 0, 1, 0)),
.Names = c("ID", "eventA", "eventB", "t1", "t2", "censored"),
class = "data.frame", row.names = c(NA, -6L))
dat$event <- with(dat, ifelse(eventA, "Censored", "Death"))
dat$id.ordered <- factor(x = dat$ID, levels = order(dat$t2, decreasing = T))
ggplot(dat, aes(x = id.ordered)) +
geom_linerange(aes(ymin = 0, ymax = t1)) +
geom_linerange(aes(ymin = t1, ymax = t2,
linetype = as.factor(censored))) +
geom_point(aes(y = ifelse(censored,
t1 + (t2 - t1) / 2, t2),
shape = event), size = 4) +
coord_flip() +
scale_linetype_manual(name = "Censoring", values = c(1, 2),
labels = c("Not censored", "Interval censored")) +
scale_shape_manual(name = "Event", values = c(19, 15)) +
# ggtitle("Interval Censoring") +
xlab("Eggs") + ylab("Weeks") +
theme_bw()CI Birds Right Censoring
It is the end of the study, and their are 5 more eggs that have not hatched yet!
Additionally, a colleague has informed you that some eggs were eaten by reptiles through out the study. They obtained the day they were eaten or lost.
Right Censoring
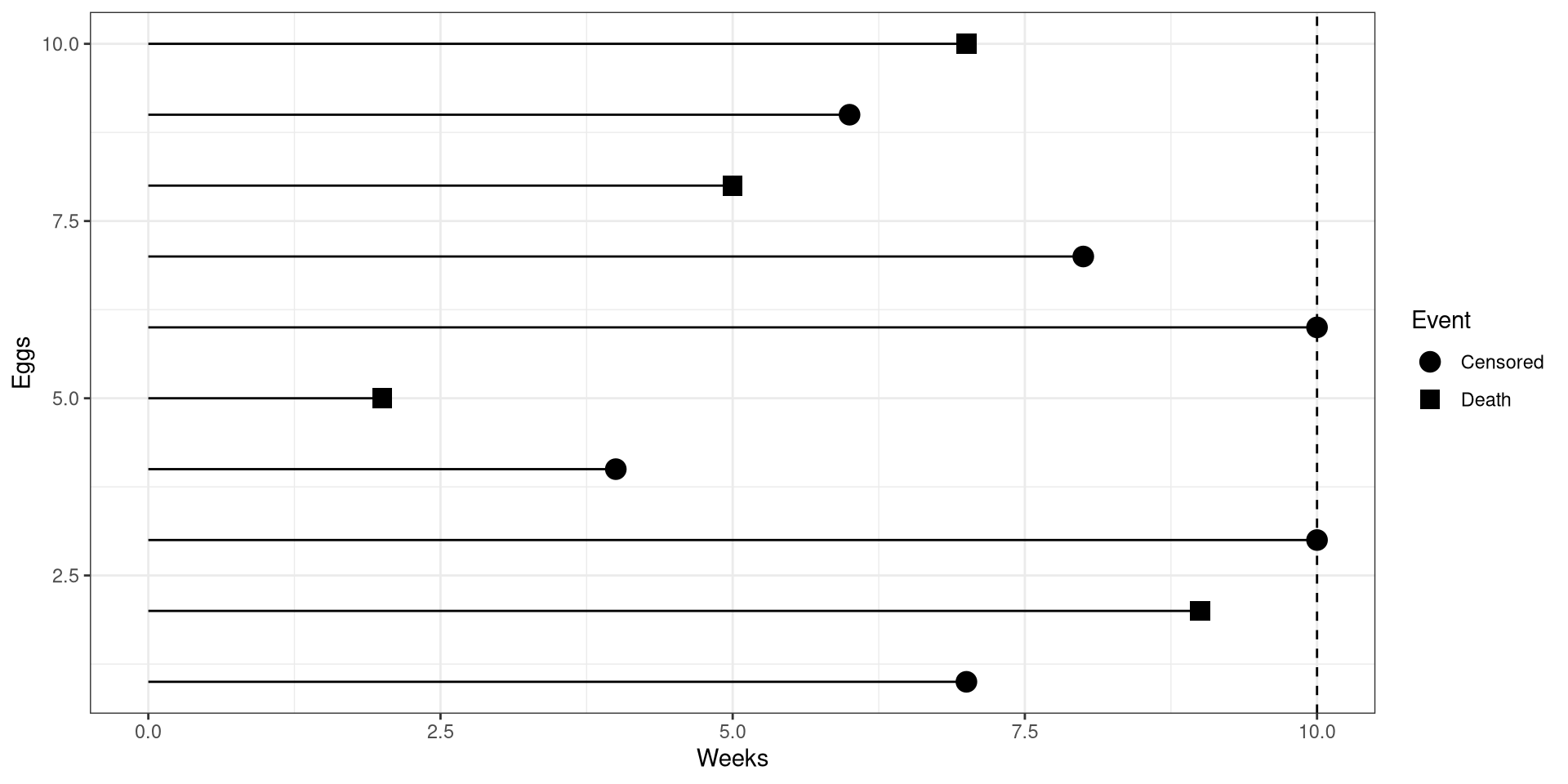
library(ggplot2)
dat <- data.frame(ID = 1:10,
t1 = c(7, 9, 10, 4, 2, 10, 8, 5, 6, 7) ,
censored = c(0, 1, 0, 0, 1, 0, 0, 1, 0, 1))
ggplot(dat, aes(x = ID, y = t1,
shape = ifelse(censored, "Death", "Censored"))) +
geom_point(size = 4) +
geom_linerange(aes(ymin = 0, ymax = t1)) +
geom_hline(yintercept = 10, lty = 2) +
coord_flip() +
scale_shape_manual(name = "Event", values = c(19, 15)) +
# ggtitle("Right Censoring") +
xlab("Patient ID") + ylab("Months") +
theme_bw()Censoring
Censoring affects the time-to-event information
However, we obtain some information when data is censored
Incorporate methods to utilize partial information
Censoring is independent of time-to-event generation
Survival Rate
Background Information
Scenarios
Censoring
Survival Rate
Example
Survival Curve
The survival curve will determine what is the probability of surviving up to a certain time
A survival curve accounts for both censored and uncensored data
A survival curve can be used to determine the median survival time of a disease
- Or the time for at least half of the eggs to hatch
Data Notation
Let \(\{t_j,d_j,R_j\}^D_{j=1}\) denote the survival data, where \(t_1<t_2<\cdots<t_D\) are the ordered distinct observed event times, \(d_j\) represents the number of events at time point \(t_j\), and \(R_j\) denotes the number of subjects still at risk of experiencing the event at \(t_j\).
Kaplan-Meier Estimator
\[ \hat{S}(t) =\left\{\begin{array}{cc} 1 & t=0 \\ \prod_{i:t_j \le t} \left( 1 - \frac{d_j}{R_j} \right) & t_j < t \end{array}\right. \]
Standard Error
\[ \widehat{SE}\{\hat S(t)\}=\sqrt{\hat S^2(t)\sum_{t_j\leq t}\frac{d_j}{R_j(R_j-d_j)}}. \]
Survival Curve
Example
Background Information
Scenarios
Censoring
Survival Rate
Example
Data
Fitting Curve
The survival package contains the necessary functions to fit a model:
Surv: Creates an outcome variablesurvfit: Fits the survival function
Plotting the Survival Function
The ggsurvfit package provides a set of tools to plot the survival function in a ggplot format. The primary functions are:
ggsurvfit: Plots Survival Functionadd_quantile: Adds line on the percentileadd_confidence_interval: Adds pointwise error bars to the plot.
Fitting Curve By Groups
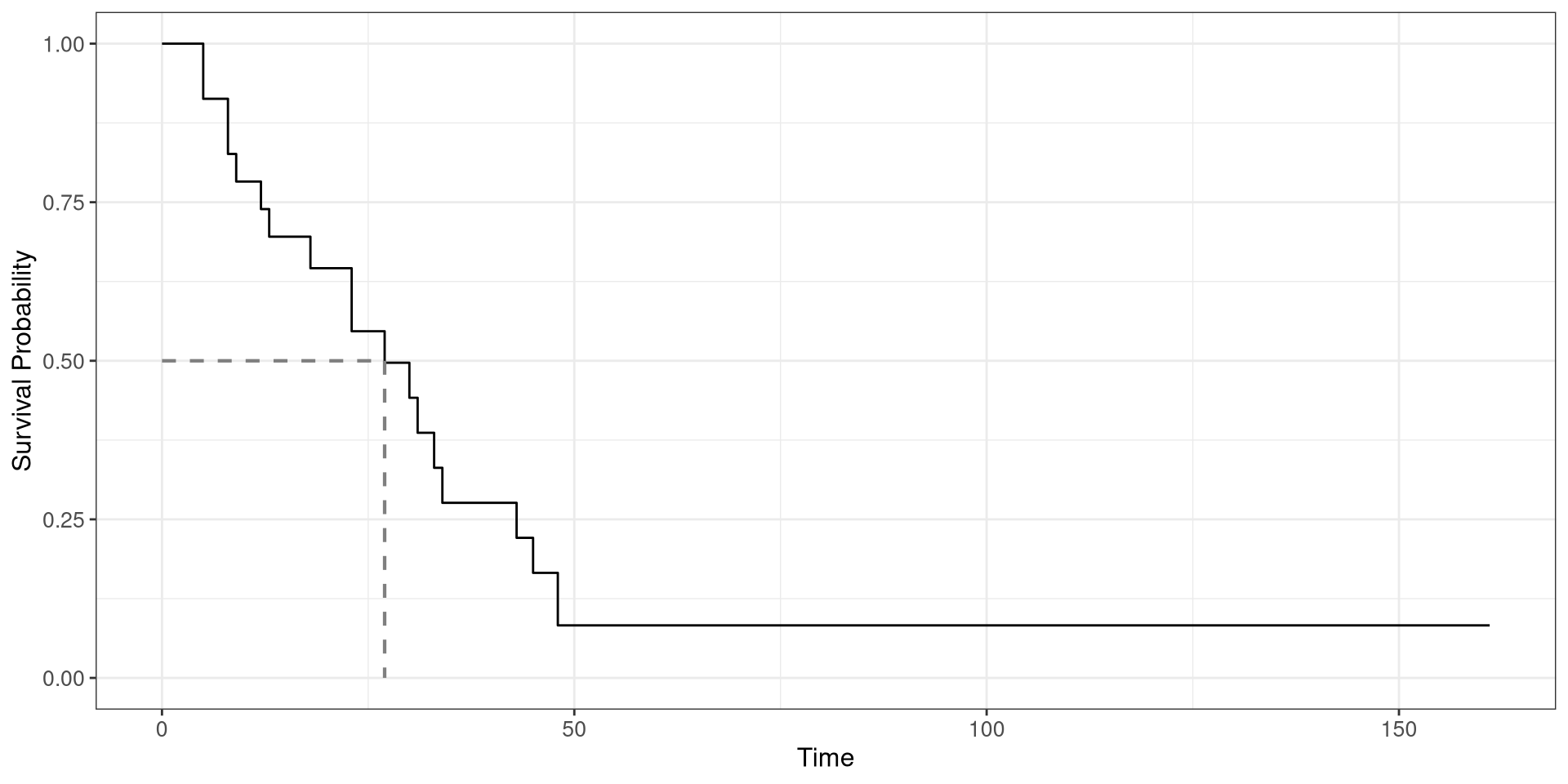
Colon Survival Curve
- Fit a survival curve using
df_colonfrom theggsurvfitpackage. time: time-to-deathstatus: censoring status- Plot the survival curve and add a line indicating the 50th percentile
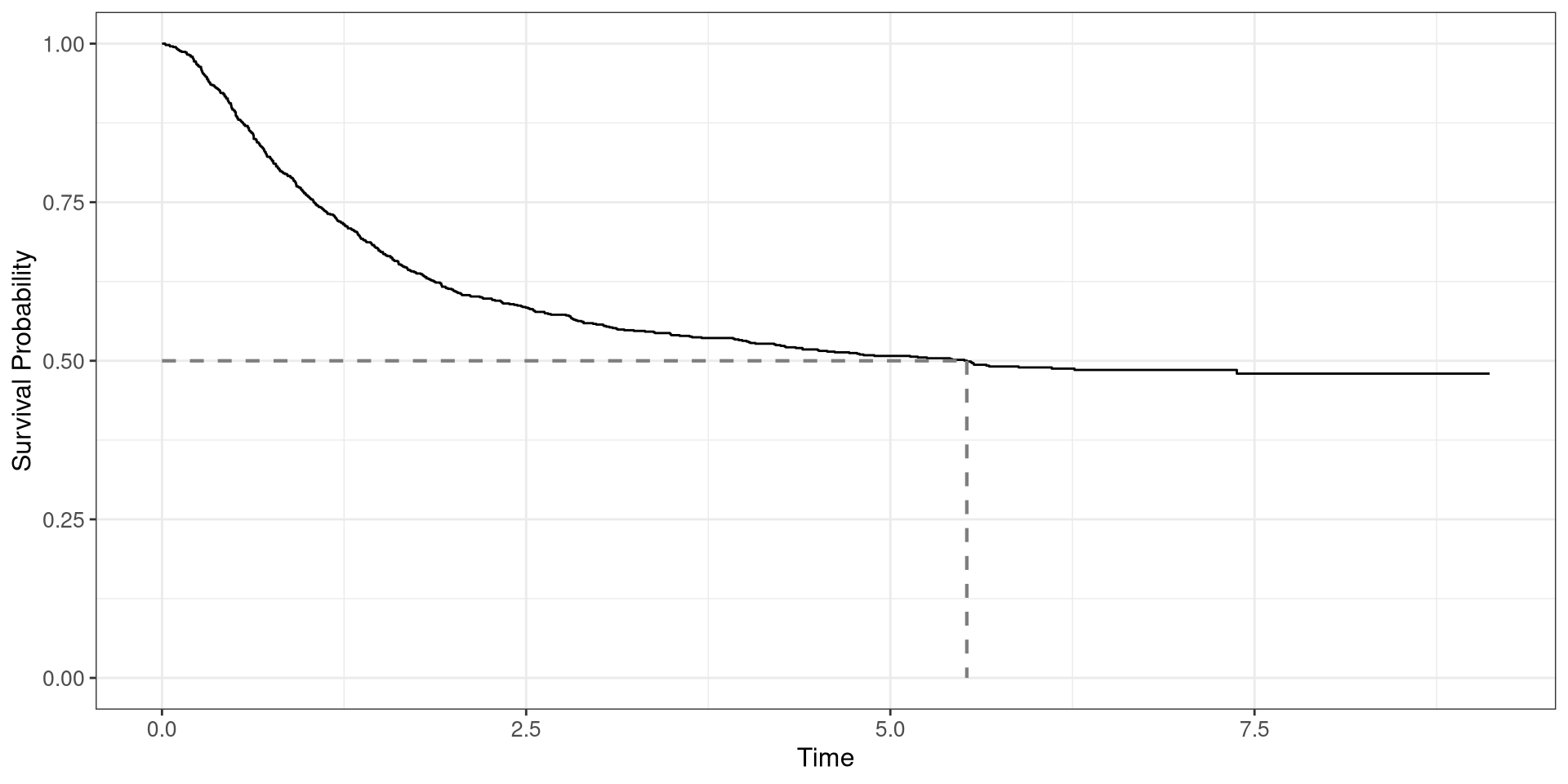
Colon Survival Curve
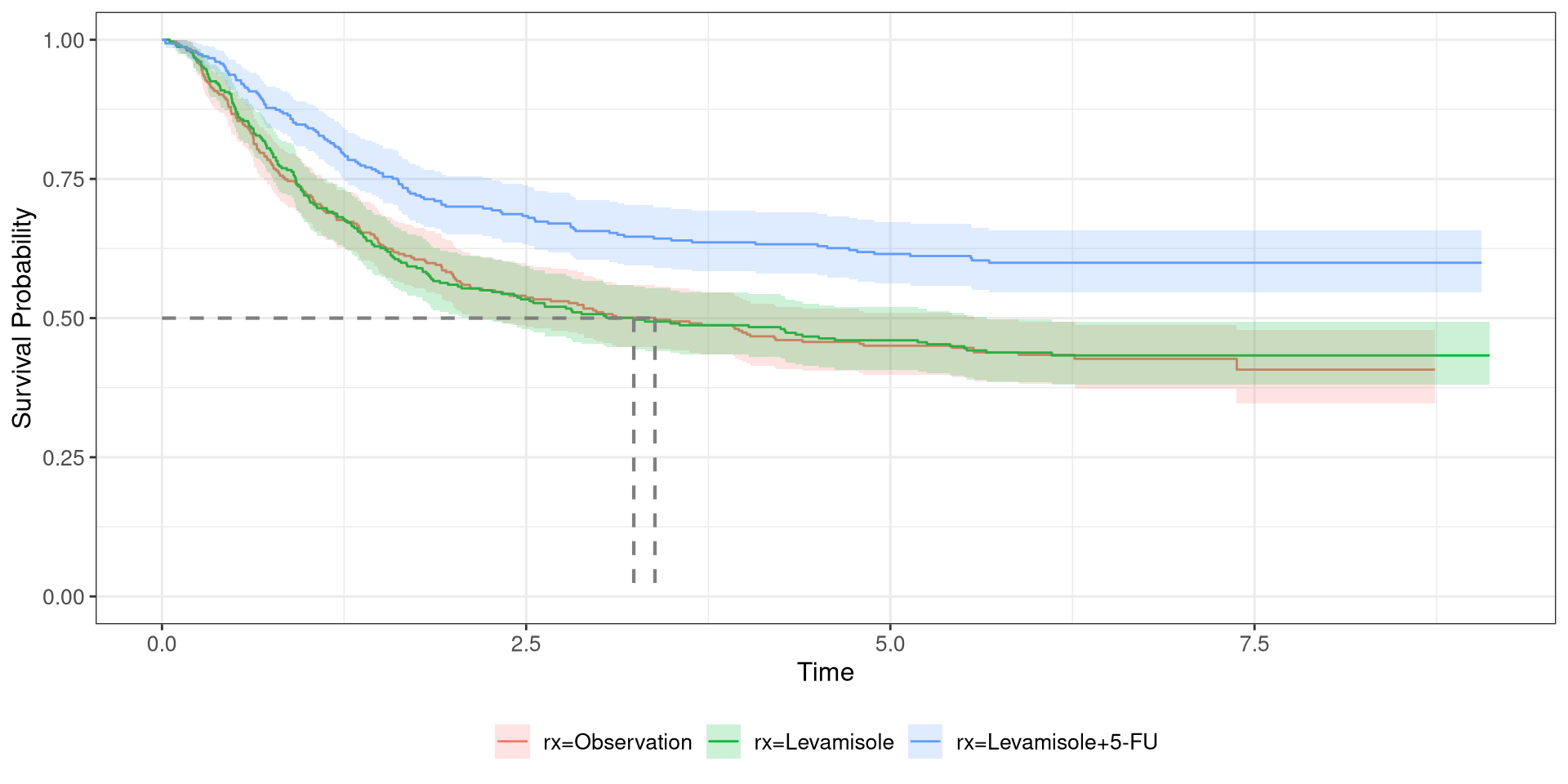
- Fit a survival curve using
df_colon, stratified by treatment regimen time: time-to-deathstatus: censoring statusrx: treatment regimen- Plot the curvival curve and add a line indicating the 50th percentile
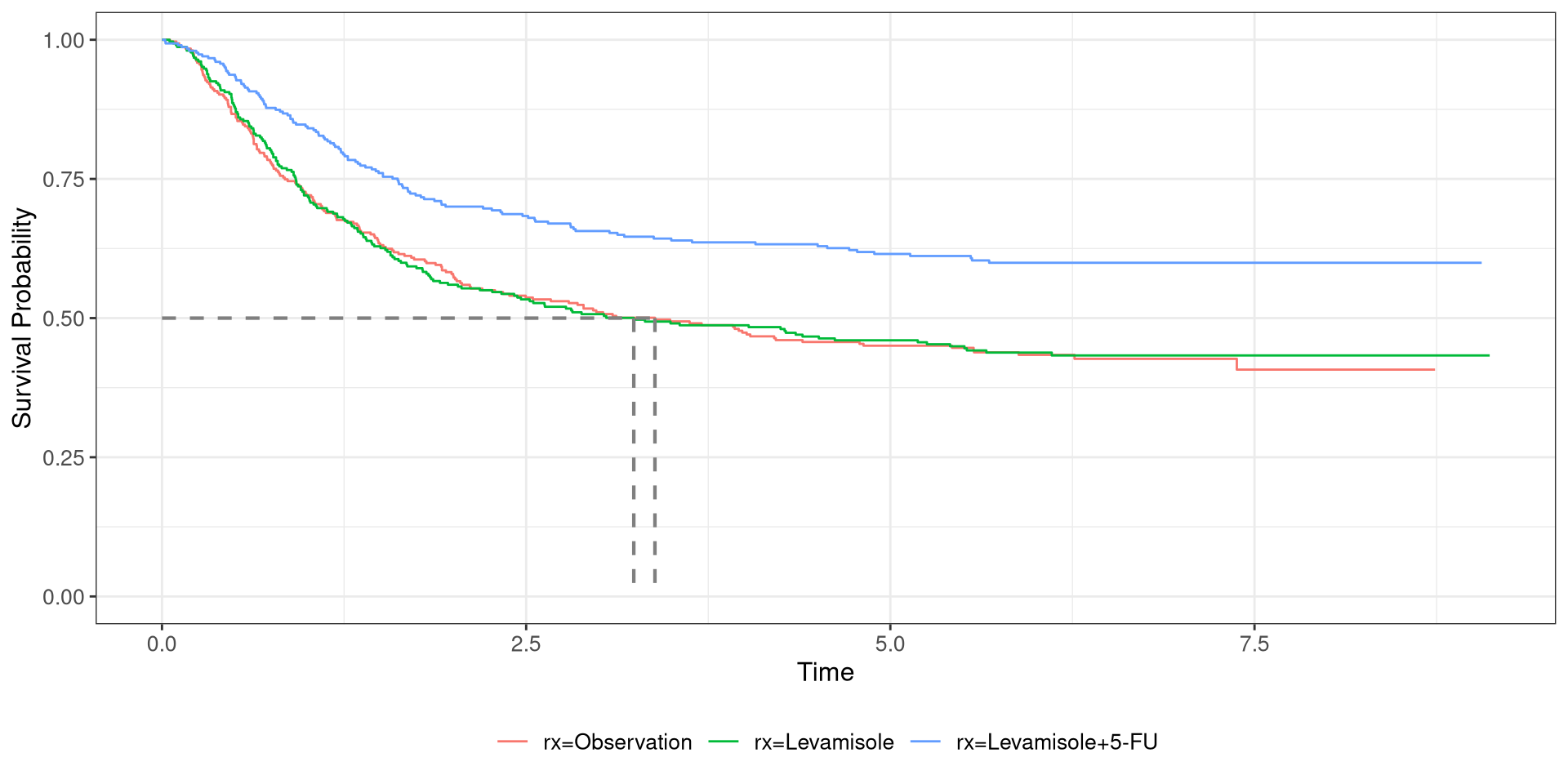
Hypothesis Testing
- When stratifying the data, Levamisole+5 FU performed better than the other regimens
- Is this a significant improvement? Or due to random chance?
- Add Confidence Intervals to see if there is a difference